I wrote a small script to generate the 3D structures of the amino acids because I wan't to investigate how well the MMFF94 force field can calculate the charge of them. I've done this before for entire proteins, but here I just want to do it one amino acid at a time. To challenge myself, I cannot rely on the excellent (o)babel tool of the Open Babel package when generating the structures. All code is on github, but have shared it on the blog too via select gists.
My SMILES for the amino acids are saved in a dictionary:
and to simply generate the structures I've updated my obutil library with the following utility functions
Finally, to generate the structures I simply run the following code
I have a separate script to calculate the charge of a molecule
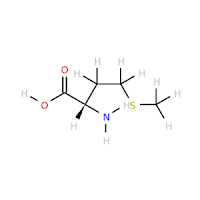
The table below is the result of the above. The images are generated using the babel tool with the -opng option.
In this case, all charges are zero on all amino acids. Notice that I did not make use of Open Babel trying to assign Hydrogen for a specific pH as that is not what I want to test. Instead, notice in the aminoacids.py file listed above there are specific patterns for obtaining (de)protonated states of a few select amino acids: ASP, GLU, HIS, LYS and ARG.
Running the (de)protonated molecules above gives the following table
The disappointing thing here is that the protonated Arginine is incorrectly assigned a charge of 0 (zero). Edit: As noted by Jan, the depiction of the protonated ARG, ASP and GLU is also wrong.
Edit2: It appears the problem is also present in the SVG depicter.
Maybe it's time to submit a bug-report?
Running the (de)protonated molecules above gives the following table
| Amino Acid | SMILES | Charge | Depiction |
|---|---|---|---|
| ASP- | NC(CC(=O)[O-])C(=O)O | -1 | |
| GLU- | NC(CCC(=O)[O-])C(=O)O | -1 | |
| HIS+ | NC(C(=O)O)Cc1c[nH+]cn1 | +1 | |
| LYS+ | NC(CCCC[NH3+])C(=O)O | +1 | |
| ARG+ | NC(CCCNC(=[NH2+])N)C(=O)O | 0 |
The disappointing thing here is that the protonated Arginine is incorrectly assigned a charge of 0 (zero). Edit: As noted by Jan, the depiction of the protonated ARG, ASP and GLU is also wrong.
Edit2: It appears the problem is also present in the SVG depicter.
Maybe it's time to submit a bug-report?